HOGENOM is a phylogenomic database providing families of homologous genes and associated phylogenetic trees (and sequence alignments) for a wide set sequenced organisms.
HOGENOM organisation
HOGENOM is divided into 13 phylum-specific databases and 1 core database. The core database contains a set of representative genomes. Each phylum-specific database contains sequences from all the organisms of the phylum plus the sequences from all the representative organims. Phylogenetic trees are build in order to present sequences from the core genome and from one of the phylum-specific database. Thus each sequence of the database can be displayed in a tree which present sequences from closely related species (sequences from the associated phylum) and sequences from a wide set of distant species (sequences from the core genome), allowing to view the sequence in both local and global evolutionnary context.
HOGENOM clusters and associated phylogenetic trees
All the proteins of the database have been clustered, then a sequences alignment was computed for each cluster.
Each alignment has been splitted into sub-alignement according to the species: 1 sub-alignment containing sequences from the core genomes, several sub-alignements containing sequences from the different phylum-specific databases.
When the "core" sub-alignment presented more than 3 sequences, a "core tree" was calculated.
Each phyla-specific sub-alignment was merged with the "core" sub-alignment, then a "phylum-specific tree" was calculated, using the "core tree" as a constraint if it exists. Thus most of "phylum-specific trees" present a common sub-tree.
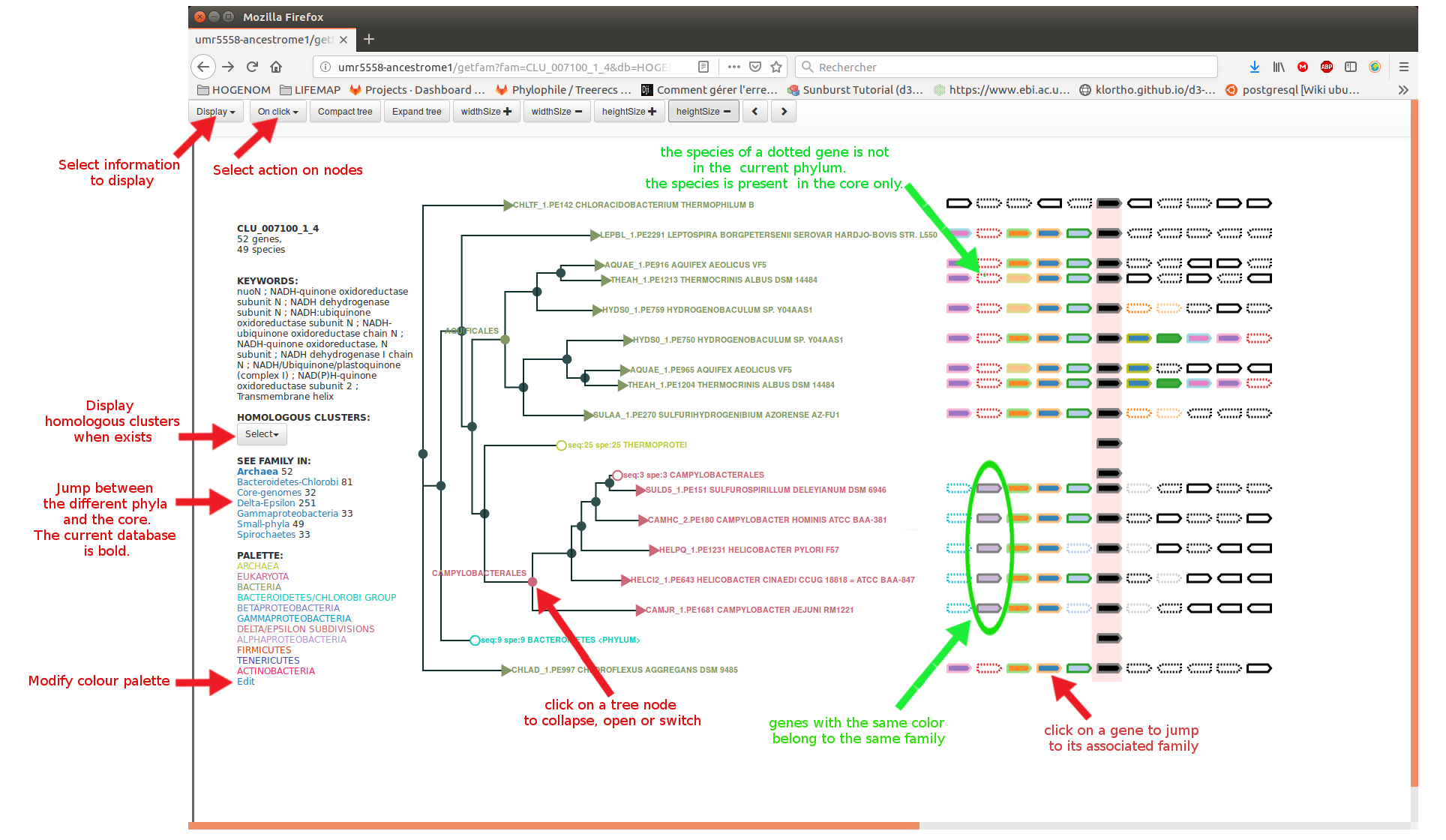
The tree display interface allows to jump between the "core tree" and the different "phylum-specific trees"
More details on the pipeline here.
HOGENOM interface